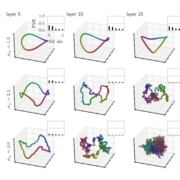
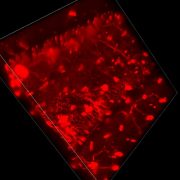
26.10.2017. Today, Prof. Kurt Zatloukal and his group together with us and the digital pathology team of 3DHISTECH, our industrial partner, completed the installation of the new generation panoramic P1000 scanner [0]. The world’s fastest whole slide image scanner (WSI) is now located in Graz. The current scanner outperforms current state-of-the-art systems by a factor 6, which provides enormous opportunities for our machine learning/AI MAKEpatho project.
Digital Pathology and Artificial Intelligence/Machine Learning
Digital pathology [1] is not just the transformation of the classical microscopic analysis of histopathological slides by pathologists to a digital visualization. Digital Pathology is an innovation that will dramatically change medical workflows in the coming years. In the center is Whole Slide Imaging (WSI), but the true added value will result from a combination of heterogenous data sources. This will generate a new kind of information not yet available today. Much information is hidden in arbitrarily high dimensional spaces and not accessible to a human pathologist. Consequently, we need novel approaches from artificial intelligence (AI) and machine learning (ML) (see definition) for exploiting the full possibilities of Digital Pathology [2]. The goal is to gain knowledge from this information, which is not yet available and not exploited to date [3].
Digital Pathology chances
Major changes enabled by digital pathology include the improvement of medical decision making with new human-AI interfaces, new chances for education and research, and the globalization of diagnostic services. The latter allows bringing the top-level expertise essentially to any patient in the world by the use of the Internet/Web. This will also generate totally new business models for worldwide diagnostic services. Furthermore, by using AI/ML we can make new information of images accessible and quantifiable (e.g. through geometrical approaches and machine learning), which is not yet available in current diagnostics. Another effect will be that digital pathology and machine learning will change the education and training systems, which will be an urgently needed solution to address the global shortage of medical specialists. While the digitalization is called Pathology 2.0 [4] we envision a Pathology 4.0 – and here explainable-AI will become important.
3DHISTECH
3DHISTECH Ltd. (the name is derived from „Three-dimensional Histological Technologies”) is a leading company, developing high-performance hardware and software products for digital pathology since 1996. As the first European manufacturer, 3DHISTECH is one of the market leaders in the world with more than 1500 sold systems. Founded by Dr. Bela MOLNAR from Semmelweis University Budapest, they are pioneers in this field, and develop high speed digital slide scanners that create high quality bright field and fluorescent digital slides, digital histology software and tissue microarray machinery. 3DHISTECH’s aim is to fully digitalize the traditional pathology workflow so that it can adapt to the ever growing demands of healthcare today. Furthermore, educational programs are also organized to help pathologists learn and master these new technologies easier.
[0] P1000 https://www.youtube.com/watch?v=WuCXkTpy5js (1:41 min)
[1] Shaimaa Al‐Janabi, Andre Huisman & Paul J. Van Diest (2012). Digital pathology: current status and future perspectives. Histopathology, 61, (1), 1-9, doi:10.1111/j.1365-2559.2011.03814.x.
[2] Anant Madabhushi & George Lee (2016). Image analysis and machine learning in digital pathology: Challenges and opportunities. Medical Image Analysis, 33, 170-175, doi:10.1016/j.media.2016.06.037.
[3] Andreas Holzinger, Bernd Malle, Peter Kieseberg, Peter M. Roth, Heimo Müller, Robert Reihs & Kurt Zatloukal (2017). Machine Learning and Knowledge Extraction in Digital Pathology needs an integrative approach. In: Springer Lecture Notes in Artificial Intelligence Volume LNAI 10344. Cham: Springer International, pp. 13-50. 10.1007/978-3-319-69775-8_2 [pdf-preprint available here]
[4] Nikolas Stathonikos, Mitko Veta, André Huisman & Paul J Van Diest (2013). Going fully digital: Perspective of a Dutch academic pathology lab. Journal of pathology informatics, 4. doi: 10.4103/2153-3539.114206
[5] Francesca Demichelis, Mattia Barbareschi, P Dalla Palma & S Forti 2002. The virtual case: a new method to completely digitize cytological and histological slides. Virchows Archiv, 441, (2), 159-164. https://doi.org/10.1007/s00428-001-0561-1
[6] Marcus Bloice, Klaus-Martin Simonic & Andreas Holzinger 2013. On the usage of health records for the design of virtual patients: a systematic review. BMC Medical Informatics and Decision Making, 13, (1), 103, doi:10.1186/1472-6947-13-103.
[7] https://www.3dhistech.com
[8] https://pathologie.medunigraz.at/forschung/forschungslabor-fuer-experimentelle-zellforschung-und-onkologie
Mini Glossary:
Digital Pathology = is not only the conversion of histopathological slides into a digital image (WSI) that can be uploaded to a computer for storage and viewing, but a complete new medical work procedure (from Pathology 2.0 to Pathology 4.0) – the basis is Virtual Microscopy.
Explainability = motivated due to lacking transparency of black-box approaches, which do not foster trust and acceptance of AI generally and ML specifically among end-users. Rising legal and privacy aspects, e.g. with the new European General Data Protection Regulations (which come into effect in May 2018) will make black-box approaches difficult to use, because they often are not able to explain why a decision has been made (see explainable AI).
Explainable AI = raising legal and ethical aspects make it mandatory to enable a human to understand why a machine decision has been made, i.e. to make machine decisions re-traceable and to explain why a decision has been made [see Wikipedia on Explainable Artificial Intelligence] (Note: that does not mean that it is always necessary to explain everything and all – but to be able to explain it if necessary – e.g. for general understanding, for teaching, for learning, for research – or in court!)
Machine Aided Pathology = is the management, discovery and extraction of knowledge from a virtual case, driven by advances of digital pathology supported by feature detection and classification algorithms.
Virtual Case = the set of all histopathological slides of a case together with meta data from the macro pathological diagnosis [5]
Virtual microscopy = not only viewing of slides on a computer screen over a network, it can be enhanced by supporting the pathologist with equivalent optical resolution and magnification of a microscope whilst changing the magnification; machine learning and ai methods can help to extract new knowlege out of the image data
Virtual Patient = has very different definitions (see [6]), we define it as a model of electronic records (images, reports, *omics) for studying e.g. diseases.
WSI = Whole Slide Image, a.k.a. virtual slide, is a digitized histopathology glass slide that has been created on a slide scanner and represents a high-resolution volume data cube which can be handled via a virtual microscope and most of all where methods from artificial intelligence generally, and interactive machine learning specifically, together with methods from topological data analysis, can make information accessible to a human pathologists, which would otherwise be hidden.
WSS = Whole Slide Scanner is the machinery for taking WSI including the hardware and the software for creating a WSI.