Call for Papers: Open Data for Discovery Science (due to July, 31, 2017)
/in Calls for Papers, HCI-KDD Events/by Andreas HolzingerThe Journal BMC Medical Informatics and Decision Making (SCI IF (2015): 2,042)
invites to submit to a new thematic series on open data for discovery science
https://bmcmedinformdecismak.biomedcentral.com/articles/collections/odds
Note: Excellent submissions to the IFIP Cross Domain Conference on Machine Learning and Knowledge Discovery (CD-MAKE), (Submission due to May, 15, 2017) relevant to the topics described below, will be invited to expand their work into this thematic series:
The use of open data for discovery science has gained much attention recently as its full potential is unfolding and being explored in projects spanning all areas of healthcare research. A plethora of data sets are now available thanks to drives to make data universally accessible and usable for discovery science. However, with these advances come inherent challenges with the processing and management of ever expanding data sources. The computational and informatics tools and methods currently used in most investigational settings are often labor intensive and rely upon technologies that have not been designed to scale and support reasoning across multi-dimensional data resources. In addition, there are many challenges associated with the storage and responsible use of open data, particularly medical data, such as privacy, data protection, safety, information security and fair use of the data. There are therefore significant demands from the research community for the development of data management and analytic tools supporting heterogeneous analytic workflows and open data sources. Effective anonymisation tools are also of paramount importance to protect data security whilst preserving the usability of the data.
The purpose of this thematic series is to bring together articles reporting advances in the use of open data including the following:
- The development of tools and methods targeting the reproducible and rigorous use of open data for discovery science, including but not limited to: syntactic and semantic standards, platforms for data sharing and discovery, and computational workflow orchestration technologies that enable the creation of data analytics, machine learning and knowledge extraction pipelines.
- Practical approaches for the automated and/or semi-automated harmonization, integration, analysis, and presentation of data products to enable hypothesis discovery or testing.
- Theoretical and practical approaches for solutions to make use of interactive machine learning to put a human-in-the-loop, answering questions including: could human intelligence lead to general heuristics that we can use to improve heuristics?
- Frameworks for the application of open data in hypothesis generation and testing in projects spanning translational, clinical, and population health research.
- Applied studies that demonstrate the value of using open data either as a primary or as an enriching source of information for the purposes of hypothesis generation/testing or for data-driven decision making in the research, clinical, and/or population health environments.
- Privacy preserving machine learning and knowledge extraction algorithms that can enable the sharing of previously “privileged” data types as open data.
- Evaluation and benchmarking methodologies, methods and tools that can be used to demonstrate the impact of results generated through the primary or secondary use of open data.
- Socio-cultural, usability, acceptance, ethical and policy issues and frameworks relevant to the sharing, use, and dissemination of information and knowledge derived from the analysis of open data.
Submission is open to everyone, and all submitted manuscripts will be peer-reviewed through the standard BMC Medical Informatics and Decision Making review process. Manuscripts should be formatted according to the submission guidelines and submitted via the online submission system. Please indicate clearly in the covering letter that the manuscript is to be considered for the ‘Open data for discovery science’ collection. The deadline for submissions will be 31 July 2017.
For further information, please email the editors of the thematic series:
Andreas HOLZINGER a.holzinger@human-centered.ai,
Philip PAYNE prpayne@wustl.edu ,or the BMC in-house editor
Emma COOKSON at emma.cookson@biomedcentral.com
Link to the IFIP Cross-Domain Conference on Machine Learning and Knowledge Extraction (CD-MAKE):
https://cd-make.net
Integrated interactomes and pathways in precision medicine by Igor Jurisica, Toronto
/in HCI-KDD Events, Lectures/by Andreas HolzingerMachine learning is the fastest growing field in computer science, and Health Informatics is amongst the greatest application challenges, providing benefits in improved medical diagnoses, disease analyses, and pharmaceutical development – towards future precision medicine.
Talk announcement: Friday, 12th May, 2017, 10:00, Seminaraum 137, Parterre, Inffeldgasse 16c
Integrated interactomes and pathways in precision medicine
by Igor Jurisica, University of Toronto and Princess Margaret Cancer Center Toronto
Abstract: Fathoming cancer and other complex disease development processes requires systematically integrating diverse types of information, including multiple high-throughput datasets and diverse annotations. This comprehensive and integrative analysis will lead to data-driven precision medicine, and in turn will help us to develop new hypotheses, and answer complex questions such as what factors cause disease; which patients are at high risk; will patients respond to a given treatment; how to rationally select a combination therapy to individual patient, etc.
Thousands of potentially important proteins remain poorly characterized. Computational biology methods, including machine learning, knowledge extraction, data mining and visualization, can help to fill this gap with accurate predictions, making disease modeling more comprehensive. Intertwining computational prediction and modeling with biological experiments will lead to more useful findings faster and more economically.
Short Bio: Igor Jurisica is Tier I Canada Research Chair in Integrative Cancer Informatics, Senior Scientist at Princess Margaret Cancer Centre, Professor at University of Toronto and Visiting Scientist at IBM CAS. He is also an Adjunct Professor at the School of Computing, Pathology and Molecular Medicine at Queen’s University, Computer Science at York University, scientist at the Institute of Neuroimmunology, Slovak Academy of Sciences and an Honorary Professor at Shanghai Jiao Tong University in China. Since 2015, he has also served as Chief Scientist at the Creative Destruction Lab, Rotman School of Management. Igor has published extensively on data mining, visualization and cancer informatics, including multiple papers in Science, Nature, Nature Medicine, Nature Methods, Journal of Clinical Oncology, and received over 9,960 citations since 2012. He has been included in Thomson Reuters 2016, 2015 & 2014 list of Highly Cited Researchers, and The World’s Most Influential Scientific Minds: 2015 & 2014 Reports.
Jurisica Lab, IBM Life Sciences Discovery Center:
Canada Tier I Research Chair: https://www.chairs-chaires.gc.ca/chairholders-titulaires/profile-eng.aspx?profileId=2347
On Nutrigenomics [1]: https://www.uhn.ca/corporate/News/Pages/Igor_Jurisica_talks_nutrigenomics.aspx
[1] Nutrigenomics tries to define the causality or relationship between specific nutrients and specific nutrient regimes (diets) on human health. The underlying idea is in personalized nutrition based on the *omics background, which may help to foster personal dietrary recommendations. Ultimately, nutrigenomics will allow effective dietary-intervention strategies to recover normal homeostasis and to prevent diet-related diseases, see: Muller, M. & Kersten, S. 2003. Nutrigenomics: goals and strategies. Nature Reviews Genetics, 4, (4), 315-322.
Cross Domain Conference for Machine Learning & Knowledge Extraction
/in Calls for Papers, HCI-KDD Events/by Andreas Holzingercd-make.net
Call for Papers – due to May, 15, 2017
https://www.wikicfp.com/cfp/servlet/event.showcfp?eventid=61244©ownerid=17803
Call for Papers due to May, 15, 2017
International IFIP Cross Domain Conference for Machine Learning & Knowledge Extraction CD-MAKE
in Reggio di Calabria (Italy) August 29 – September 1, 2017
CD stands for Cross-Domain and means the integration and appraisal of different fields and application domains (e.g. Health, Industry 4.0, etc.) to provide an atmosphere to foster different perspectives and opinions. The conference is dedicated to offer an international platform for novel ideas and a fresh look on the methodologies to put crazy ideas into Business for the benefit of the human. Serendipity is a desired effect, and shall cross-fertilize methodologies and transfer of algorithmic developments.
MAKE stands for MAchine Learning & Knowledge Extraction.
CD-MAKE is a joint effort of IFIP TC 5, IFIP WG 8.4, IFIP WG 8.9 and IFIP WG 12.9 and is held in conjunction with the International Conference on Availability, Reliability and Security (ARES).
Keynote Speakers are Neil D. LAWRENCE (Amazon) and Marta MILO (University of Sheffield).
IFIP is the International Federation for Information Processing and the leading multi-national, non-governmental, apolitical organization in Information & Communications Technologies and Computer Sciences, is recognized by the United Nations and was established in the year 1960 under the auspices of the UNESCO as an outcome of the first World Computer Congress held in Paris in 1959.
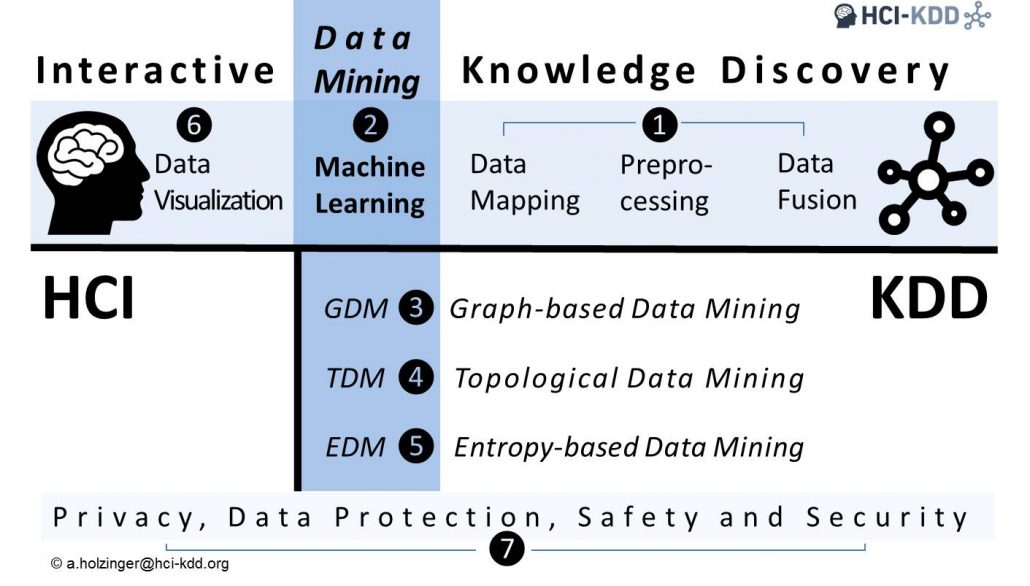
Papers are sought from the following seven topical areas (see image below). Papers which deal with fundamental questions and theoretical aspects in machine learning are very welcome.
❶ Data science (data fusion, preprocessing, data mapping, knowledge representation),
❷ Machine learning (both automatic ML and interactive ML with the human-in-the-loop),
❸ Graphs/network science (i.e. graph-based data mining),
❹ Topological data analysis (i.e. topology data mining),
❺ Time/entropy (i.e. entropy-based data mining),
❻ Data visualization (i.e. visual analytics), and last but not least
❼ Privacy, data protection, safety and security (i.e. privacy aware machine learning).
Proposals for Workshops, Special Sessions, Tutorials: April, 19, 2017
Submission Deadline: May, 15, 2017
Author Notification: June, 14, 2017
Camera Ready Deadline: July, 07, 2017
https://cd-make.net/call-for-papers
Call for Papers – Privacy Aware Machine Learning PAML due to April, 1, 2017
/in Calls for Papers, HCI-KDD Events/by Andreas HolzingerPrivacy Aware Machine Learning (PAML)
for Health Data Science
Special Session on September, 1, 2017, organized by Andreas HOLZINGER, Peter KIESEBERG, Edgar WEIPPL and A Min TJOA in the context of the 12th International Conference on Availability, Reliability and Security (ARES and CD-ARES), Reggio di Calabria, Italy, August 29 – September, 2, 2017
supported by the International Federation of Information Processing IFIP > TC5 and WG 8.4 and WG 8.9
https://cd-ares-conference.eu
https://www.ares-conference.eu
Keynote Talk by Neil D. LAWRENCE, University of Sheffield and Amazon
With the new European data protection and privacy regulations coming into effect with January, 1, 2018 issues having been nice to have so far are becoming a must have. Privacy aware machine learning will be one of the most important fields for the European research community and the IT business in particular. Most affected is the whole area of biology, medicine and health, partiuclarly driven by the fact that health sciences are becoming a more and more data intensive science.
This special session will bring together scientists with diverse background, interested in both the underlying theoretical principles as well as the application of such methods for practical use in the biomedical, life sciences and health care domain. The cross-domain integration and appraisal of different fields will provide an atmosphere to foster different perspectives and opinions; it will offer a platform for novel crazy ideas and a fresh look on the methodologies to put these ideas into business.
All paper will be peer-reviewed by three members of the international PAML-commitee. Paper acceptance rate of the last session was 35 %. Accepted papers will be published in a Springer Lecture Notes in Computer Science (LNCS) Volume and excellent contributions will be invited to be extented in a special issue of a journal (planned Springer MACH and/or BMC MIDM).
Research topics covered by this special session include but are not limited to the following topics:
– Production of Open Data Sets
– Synthetic data sets for learning algorithm testing
– Privacy preserving machine learning, data mining and knowledge discovery
– Data leak detection
– Data citation
– Differential privacy
– Anonymization and pseudonymization
– Securing expert-in-the-loop machine learning systems
– Evaluation and benchmarking
This picture was taken by our local host, Francesco Buccafurri on January, 3, 2017: from the conference venue you have a direct view to the Aetna volcano:
Papers due to April, 30, 2016: Privacy Aware Machine Learning (PAML) for Health Data Science
/in Calls for Papers, HCI-KDD Events/by Andreas HolzingerWe are organizing a special session on Privacy Aware Machine Learning for Health Data Science at the 11th international Conference on Availability, Reliability and Security (ARES and CD-ARES), Salzburg, Austria, August 29 – September, 2, 2016
supported by the International Federation of Information Processing IFIP > TC5 and WG 8.4 and WG 8.9
https://cd-ares-conference.eu
https://www.ares-conference.eu
Keynote Talk by Bernhard SCHÖLKOPF, Max Planck Institute for Intelligent Systems, Empirical Inference Department

We are proud to welcome Bernhard Schölkopf as Keynote Speaker to the ARES/CD-ARES conference in Salzburg
Machine learning is the fastest growing field in computer science [Jordan, M. I. & Mitchell, T. M. 2015. Machine learning: Trends, perspectives, and prospects. Science, 349, (6245), 255-260], and it is well accepted that health informatics is amongst the greatest challenges [LeCun, Y., Bengio, Y. & Hinton, G. 2015. Deep learning. Nature, 521, (7553), 436-444 ], e.g. large-scale aggregate analyses of anonymized data can yield valuable insights addressing public health challenges and provide new avenues for scientific discovery [Horvitz, E. & Mulligan, D. 2015. Data, privacy, and the greater good. Science, 349, (6245), 253-255]. Privacy is becoming a major concern for machine learning tasks, which often operate on personal and sensitive data. Consequently, privacy, data protection, safety, information security and fair use of data is of utmost importance for health data science.
The amount of patient-related data produced in today’s clinical setting poses many challenges with respect to collection, storage and responsible use. For example, in research and public health care analysis, data must be anonymized before transfer, for which the k-anonymity measure was introduced and successively enhanced by further criteria. As k-anonymity is an NP-hard problem, which cannot be solved by automatic machine learning (aML) approaches we must often make use of approximation and heuristics. As data security is not guranteed given a certain k-anonymity degree, additional measures have been introduced in order to refine results (l-diversity, t-closeness, delta-presence). This motivates methods, methodologies and algorithmic machine learning approaches to tackle the problem. As the resulting data set will be a tradeoff between utility, usability and individual privacy and security, we need to optimize those measures to individual (subjective) standards. Moreover, the efficacy of an algorithm strongly depends on the background knowledge of an potential attacker as well as the underlying problem domain. One possible solution is to make use of interactive machine learning (iML) approaches and put a human-in-the-loop where the central question remains open: “could human intelligence lead to general heuristics we can use to improve heuristics?”
Research topics covered by this special session include but are not limited to the following topics:
– Production of Open Data Sets
– Synthetic data sets for learning algorithm testing
– Privacy preserving machine learning, data mining and knowledge discovery
– Data leak detection
– Data citation
– Differential privacy
– Anonymization and pseudonymization
– Securing expert-in-the-loop machine learning systems
– Evaluation and benchmarking
This special session will bring together scientists with diverse background, interested in both the underlying theoretical principles as well as the application of such methods for practical use in the biomedical, life sciences and health care domain. The cross-domain integration and appraisal of different fields will provide an atmosphere to foster different perspectives and opinions; it will offer a platform for novel crazy ideas and a fresh look on the methodologies to put these ideas into business.
Accepted Papers will be published in a Springer Lecture Notes in Computer Science LNCS Volume.
Schedule:
I) Deadline for submissions: April, 30, 2016
Paper submission via:
https://cd-ares-conference.eu/?page_id=43
II) Camera Ready deadline: July, 4, 2016
III) Special Session: August, 30, 2016
> Conference Venue
> Travel Information Salzburg
> Lonely Planet Salzburg
The International Scientific Committee – consisting of experts from the international expert network HCI-KDD dealing with area (7), privacy, data protection, safety and security and additionally invited international experts will ensure the highest possible scientific quality, each paper will be reviewed by at least three reviewers (the paper acceptance rate of the last special session was 35 %).
January, 26, 2016, Workshop “Machine Learning for Biomedicine” TU Graz
/in HCI-KDD Events/by Andreas HolzingerDate: Tuesday, 26th January 2016, Start: 10:00, End: 17:00; Venue: Graz University of Technology,
Institute of Computer Graphics and Knowledge Visualization CGV, hosted by Prof. Tobias SCHRECK
Address: Inffeldgasse 16c, A-8010 Graz <maps and directions>
Machine learning is the most growing field in computer science [Jordan, M. I. & Mitchell, T. M. 2015. Machine learning: Trends, perspectives, and prospects. Science, 349, (6245), 255-260], and it is well accepted that health informatics is amongst the greatest challenges [LeCun, Y., Bengio, Y. & Hinton, G. 2015. Deep learning. Nature, 521, (7553), 436-444 ].
Sucessful Machine Learning for Health Informatics requires a comprehensive understanding of the data ecosystem and a multi-disciplinary skill-set, from seven specializations: 1) data science, 2) algorithms, 3) network science, 4) graphs/topology, 5) time/entropy, 6) data visualization and visual analytics, and 7) privacy, data protection, safety and security – as supported by the international expert network HCI-KDD.
Program see: https://human-centered.ai/machine-learning-for-biomedicine-tugraz/
Workshop “Machine Learning for Health Informatics” November, 30, 2015
/in HCI-KDD Events, Lectures/by Andreas HolzingerWorkshop
Machine Learning for Health Informatics
Machine learning is a large and rapidly developing subfield of computer science that evolved from artificial intelligence (AI) and is tightly connected with data mining and knowledge discovery. The ultimate goal of machine learning is to design and develop algorithms which can learn from data. Consequently, machine learning systems learn and improve with experience over time and their trained models can be used to predict outcomes of questions based on previously seen knowledge. In fact, the process of learning intelligent behaviour from noisy examples is one of the major questions in the field. The ability to learn from noisy, high dimensional data is highly relevant for many applications in the health informatics domain. This is due to the inherent nature of biomedical data, and health will increasingly be the focus of machine learning research in the near future.
Program
https://human-centered.ai/machine-learning-for-health-informatics/
Apr, 14, 2015 Seminar Talks Deep Learning
/in HCI-KDD Events/by Andreas HolzingerTitle: Using Deep Learning for Discovering Knowledge from Images: Pitfalls and Best Practices
Lecturer: Marcus BLOICE <expertise>
Abstract: Neural networks have been shown to be adept at image analysis and image classification. Deep layered neural networks especially so. However, deep learning requires two things in order to work proficiently: large amounts of data and lots of processing power. In this talk both aspects are covered, allowing you to maximise the potential of deep learning. Firstly, we will learn how the computational power of GPUs can be used to speed up learning by orders of magnitude, making it possible to learn from very large datasets on commodity hardware. Thanks to software such as Theano, Caffe, and Pylearn2, the GPU can be leveraged without needing to be an expert in parallel programming. This talk will discuss how. Secondly, data preprocessing, data augmentation, and artificial data generation are discussed. These methods allow you to ensure you are making the most of the data you possess, by expanding your dataset and preparing your data properly before analysis. This means discussing best practices in data preparation, using methods such as histogram equalisation, contrast stretching or normalisation, and discussing artificial data generation in detail. The tools you require to do so are described, using multi-platform software that is freely available. Finally, the talk will touch on hyper-parameters and the best practices and pitfalls of hyper-parameter choice when training deep neural networks.
Title: Pitfalls for applying Machine Learning in HCI-KDD: Things to be aware of and how to avoid them
Lecturer: Christof STOCKER <expertise>
Abstract: When dealing with big and unstructured data sets, we often try to be creative and to experiment with a number of different approaches for the purpose of knowledge discovery. This can lead to new insights and even spark novel ideas. However, ignorant application of algorithms to unknown data is dangerous and can lead to false conclusions – with high statistical significance. In finite data sets, structure can emerge from sheer randomness. Furthermore, hidden variables can lead to significant correlations that in turn might result in wrong conclusions. Beyond this, data science as a discipline has developed into a complex area in which mistakes can occur with ease and even lead experienced scientists astray. In this talk we will investigate these pitfalls together on simple examples and discuss how we can address these concerns with manageable effort.
Feb, 17, 2015 > Seminar Talk by Hubert Wagner
/in HCI-KDD Events/by Andreas HolzingerTitle: Topological analysis of text data.
Lecturer: Hubert WAGNER <expertise>
Abstract: In this talk an ongoing effort will be described to apply persistent homology in the area of text data mining. Persistent homology is the main tool of topological data analysis. In essence, it allows to robustly describe the shape of a data set, and compare the shapes of different data sets.
First, persistent homology will be explained, emphasizing its intuitive side.
Then, it will be demonstrated how persistent homology can be applied in the context of analyzing sets of text documents. Using the vector space model interpretation, each document becomes a point in a high-dimensional space, and it is intuitive to ask about the shape of such a point cloud. It wil be discussed, how this information can be used for knowledge discovery. Finally, an algorithmic aspect is emphasized, which is crucial if industrial applications are to be tackled.
Biography: Hubert Wagner is a computer scientist, currently working as a Postdoc at the Institute of Science and Technology Austria (IST-Austria) at the Edelsbrunner Group. Having worked as a software engineer, he moved towards science and obtained a PhD degree in 2014 from the Jagiellonian University in Krakow, Poland. Hubert is interested in the application of computational geometry and topology and related algorithmic questions. He is convinced that tools such as persistent homology may offer novel and robust solutions to many problems he encountered as an engineer, including e.g. problems in text mining. This line of his research was supported by a Google Research Grant from 2011 to 2012 (with Prof. Marian Mrozek and Dr. Paweł Dłotko) and is now continued within the Topological Complex Systems (TOPOSYS) grant. Efficient algorithms and their implementations are an important part of his work.
More Information: https://publist.ist.ac.at/ist/people/180-Hubert_Wagner/works