Current as of: June, 21, 2019 – 21:00 CET
Machine Learning for Health Informatics
“It is remarkable that a science which began with the consideration of games of chance
should have become the most important object of human knowledge”
Pierre Simon de Laplace, 1812.
2019S, 2.0 h 3.0 ECTS, Type: VU Lecture with Excercises, Language: English
Venue: Vienna University of Technology > Faculty of Informatics
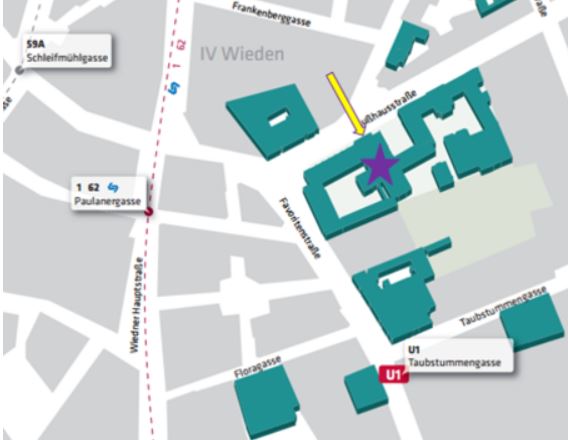
*CLASS OF 2019 STARTS: Tuesday, 12th March 2019, 17:30 – 20:30*
Seminarraum 127, Gußhausstrasse 25-29, Stiege 1, 3. Stock
>> Link to TISS
Lecturers: Andreas HOLZINGER, Holzinger Group HCI-KDD
Rudi FREUND, Theory & Logic Group
Tutors: Marcus BLOICE, Anna SARANTI, Florian ENDEL
>> Course Syllabus Class of 2019
Questions to: andreas.holzinger AT tuwien.ac.at
Introduction Paper: Machine Learning for Health Informatics (pdf, 2,000 kB – reference as: Andreas Holzinger 2016. Machine Learning for Health Informatics. In: Lecture Notes in Artificial Intelligence LNAI 9605. Springer, pp. 1-24, doi:10.1007/978-3-319-50478-0_1)
Introduction Video: https://www.youtube.com/watch?v=lc2hvuh0FwQ (Students please watch this video first)
Course Goals:
Health is increasingly turning into a data driven business. AI/ML provides the necessary methods, algorithms and tools, and the health domain provides the necessary data and domain expertise. To enable successful solutions for the benefit of the patients, health industry urgently needs a new kind of graduates!
This graduate course follows a research-based teaching (RBT) approach and discusses experimental methods for combining human intelligence with machine learning to solve problems from health informatics. The focus of the class of 2019 is even more on explainable-AI, causality and ethical, social and public issues of AI/ML for health informatics. See here a short (6 min) Youtube Video on explainable AI
For German readers: Andreas Holzinger 2018. Explainable AI (ex-AI). Informatik-Spektrum, 41, (2), 138-143, doi:10.1007/s00287-018-1102-5
For practical applications we focus on Python – which is to date the worldwide most used ML-language. Tutorial: Python-Tutorial-for-Students-Machine-Learning-course (pdf, 2,279 kB – reference as: Marcus D. Bloice & Andreas Holzinger 2016. A Tutorial on Machine Learning and Data Science Tools with Python. In: Lecture Notes in Artificial Intelligence LNAI 9605. Springer, pp. 437-483, doi:10.1007/978-3-319-50478-0_22)
Why should I study
AI/Machine Learning for Health Informatics?
1) AI/machine learning (> differences) for health is rapidly growing
AI/Machine learning (ML) is the most growing field in computer science (Jordan & Mitchell, 2015. Machine learning: Trends, perspectives, and prospects. Science, 349, (6245), 255-260), and it is well accepted that health informatics is amongst the greatest challenges (LeCun, Bengio, & Hinton, 2015. Deep learning. Nature, 521, (7553), 436-444), with Privacy Aware Machine Learning (PAML) as a must!
The future of medicine is in the data and privacy aware machine (un-)learning is no longer a nice to have, but a must.
ML is changing the future of health: Internationally outstanding universities count on the combination of machine learning and health informatics and expand these fields, for example: Carnegie-Mellon University, Harvard, Stanford – or in Europe ETH, RWTH just to name a few!
2) AI/machine learning for health informatics pose enormous Business Opportunities:
McKinsey: An executive’s guide to machine learning
NY Times: The Race Is On to Control Artificial Intelligence, and Tech’s Future
Economist: Million-dollar babies
3) AI/machine learning for health informatics provide career chances for TU graduates:
“Fei-Fei Li, a Stanford University professor who is an expert in computer vision, said one of her Ph.D. candidates had an offer for a job paying more than $1 million a year, and that was only one of four from big and small companies.”
https://www.mckinsey.com/industries/high-tech/our-insights/an-executives-guide-to-machine-learning
4) AI/machine learning for health informatics offers market opportunities for spin-offs:
“By 2020, the market for machine learning applications will reach $40 billion, IDC, a market research firm, estimates.
By 2018, IDC predicts, at least 50 percent of developers will include A.I. features in what they create.”
https://www.nytimes.com/2016/03/26/technology/the-race-is-on-to-control-artificial-intelligence-and-techs-future.html?_r=2
Description:
The goal of ML is to develop algorithms which can learn and improve over time and can be used for predictions. In automatic Machine learning (aML), great advances have been made, e.g., in speech recognition, recommender systems, or autonomous vehicles. Automatic approaches, e.g. deep learning, greatly benefit from big data with many training sets. In the health domain, sometimes we are confronted with a small number of data sets or rare events, where aML-approaches suffer of insufficient training samples. Here interactive Machine Learning (iML) may be of help, having its roots in Reinforcement Learning (RL), Preference Learning (PL) and Active Learning (AL). The term iML can be defined as algorithms that can interact with agents and can optimize their learning behaviour through these interactions, where the agents can also be human. This human-in-the-loop can be beneficial in solving computationally hard problems, e.g., subspace clustering, protein folding, or k-anonymization, where human expertise can help to reduce an exponential search space through heuristic selection of samples. Therefore, what would otherwise be an NP-hard problem reduces greatly in complexity through the input and the assistance of a human agent involved in the learning phase. However, although humans are excellent at pattern recognition in dimensions of ≤3; most biomedical data sets are in dimensions much higher than 3, making manual data analysis very hard. Successful application of machine learning in health informatics requires to consider the whole pipeline from data preprocessing to data visualization. Consequently, this course fosters the HCI-KDD approach, which encompasses a synergistic combination of methods from two areas to unravel such challenges: Human-Computer Interaction (HCI) and Knowledge Discovery/Data Mining (KDD), with the goal of supporting human intelligence with machine learning.
Grading:
Machine learning is a highly practical field, consequently this class is a VU: there will be a written exam at the end of the course, and during the course the students will solve related assignments. ECTS Breakdown: 75 hours in 15 hours lecture, 15 hours preparation for the lecture and practicals, 30 hours assignments, 15 hours preparation for the 1 hour written exam.
Course Content:
For the successful application of ML in health informatics a comprehensive understanding of the whole HCI-KDD-pipeline, ranging from the physical data ecosystem to the understanding of the end-user in the problem domain is necessary. In the medical world the inclusion of privacy, data protection, safety and security is mandatory.
Differentiation from and bridging to existing courses:
At the TU Vienna are currently the following courses on “machine learning”, i.e.
183.605 Machine Learning for Visual Computing, 3 VU, 4,5 ECTS, in winter term, which deals with linear models for regression and classification (Perceptron, Linear Basis Function Models), applications in computer vision, neural nets, error functions and optimization (e.g., pseudo-inverse, gradient descent, newton method), model complexity, regularization, model selection, Vapnik-Chernovenkis dimension, kernel methods: duality, sparsity, Support Vector Machine, principal component analysis and Hebbian rule, canonical correlation analysis, Bayesian regression, relevance vector machine, clustering und vector quantization (e.g., k-means), overview of deep learning models; the ECTS breakdown is as follows: 112,5 hours in 30 hours lecture time, 70 hours for two assignments, 2,5 hours interviews, 1 hour written exam plus 9 hours preparation time.
183.663 Deep Learning for Visual Computing, 2 VU, 3 ECTS, in winter term, covers Deep Learning for automatic image analysis, e.g. for classifying images into categories or detecting and distinguishing persons; where deep Learning has recently lead to breakthroughs; in certain problems, the performance of current methods based on this technology is similar or even better than that of humans – a novelty in this field. The goal of this lecture is to provide a comprehensive introduction to Deep Learning and its application for solving practical problems, i.e. Computer Vision and Image Processing, parametric models, iterative optimization, feedforward Neural Networks, backpropagation, convolutional Neural Networks for classification, detection, and segmentation, Software libraries and practical aspects, Preprocessing, data augmentation, regularization, visualizations, guest lectures on medical applications and ethical aspects; the ECTS breakdown is as follows: 75 hours in 16 hours lectures, 34 hours programming exercises, 24 hours exam preparation, 1 hour written exam
184.702 3VU Machine Learning, in winter term, which deals mainly with principles of supervised and unsupervised ML, including pre-processing and data preparation, as well as evaluation of Learning Systems. ML models discussed may include e.g. Decision Tree Learning, Model Selection, Bayesian Networks, Support Vector Machines, Random Forests, Hidden Markov Models, as well as ensemble methods;
Besides from focusing on health informatics (biological, biomedical, medical, clinical) and health related problems, we will build on and refer to the courses above, to avoid any parallelization, thus will particularly focus on solving problems of health with other ML-approaches (both aML and iML).
Consequently, this course is an addtional benefit for the students of computer science to foster machine learning and to show some examples in the important area of health informatics which is currently a hot topic internationally and opens a lot of future opportunities.
Lecture 01 – Week 11, Tuesday, March, 12, 2019
Module 01 – Introduction: Machine learning for health informatics: Introduction, challenges and future directions
Lecture Outline: In this first module we get a rough overview on the differences between automatic machine learnig and interactive machine learning and discuss a few future challenges of the HCAI approach to ensure ethical responsible AI/ML. This shall emphasize the integrative ML approach, where at first we learn from prior data, then extract knowledge in order to generalize and to detect certain patterns in the data and use these to make predictions and help to make decisons under uncertainty. The grand future goal is in understandability, re-traceability and explainability.
Lecture Keywords: HCI-KDD approach, integrative AI/ML, complexity, automatic ML, interactive ML, explainable AI
Topic 01: The HCI-KDD approach: towards integrative machine learning
Topic 02: Application Area Health: On the complexity of health informatics
Topic 03: Probabilistic learning on the example of Gaussian processes
Topic 04: Automatic Machine Learning (aML)
Topic 05: Interactive Machine Learning (iML)
Topic 06: Causality vs. Causability
Topic 07: Towards explainable-AI
Conclusion and Future Outlook
To get a preview you can have a look at the slides of the last course years: 2018, 2017, 2016
however, please note that for the 2019 exam of course the 2019 slides are relevant
Learning Goals: At the end of the first lecture the students …
+ become aware of some problems of the application domain medicine and health
+ have an overview on current trends, challenges, hot topics and future aspects of AI/ML for health informatics
+ know the differences, advantages and disadvantages of automatic ML and interactive ML
+ get an understanding of the importance of re-traceability, transparency, explainability and causality
Reading for Students: (some prereading/postreading and video recommendations):
- Holzinger, A. 2016. Interactive Machine Learning for Health Informatics: When do we need the human-in-the-loop? Springer Brain Informatics, 1-13. doi: doi: 10.1007/s40708-016-0042-6
- Dossier: HOLZINGER (2016) Dossier interactive Machine Learning Health Informatics
- Watch the video of Andreas Holzinger: https://youtu.be/lc2hvuh0FwQ
- Watch the video of Google DeepMindHealth: https://youtu.be/teZ2m5oTKwM
- “Medicine is so complex, the challenges are so great … we need everything that we can bring to make our diagnostics more precise, more accurate and our therapeutics more focused on that patient.” Sir Malcolm GRANT, NHS England, in: Machine learning : ROYAL SOCIETY Conference report, Part of the conference series Breakthrough science and technologies Transforming our future with machine learning), https://royalsociety.org/topics-policy/projects/machine-learning
Watch the videos: https://www.youtube.com/playlist?list=PLg7f-TkW11iX3JlGjgbM2s8E1jKSXUTsG
Lecture 02 – Week 12, Tuesday, March, 19, 2019
Module 02 – From Clinical Decision Making to explainable AI (ex-AI): selected methods of transparent machine learning
Lecture Outline: Medical action is permanent decsion making under uncertainty within limited time (“5 -Minutes”). The problem of the most successful AI/ML methods (e.g. deep learning; see the differences between AI-ML-DL here) is that they are often considered to be “black-boxes” which is not quite true. However, even if we understand the underlying mathematical and theoretical principles, it is difficult to re-enact and to answer the question of why a certain machine decision has been reached. A general serious drawback is that such models have no explicit declarative knowledge representation, hence have difficulty in generating the required explanatory structures – the context – which considerably limits the achievement of their full potential. Interestingly the “old symbolic and logic based AI-approaches” did have such explanatory structures, at least for a very narrow domain space. One future goal is in implicit knowledge elicitation through efficient human-AI interfaces.
Lecture Keywords: clinical decsion making, transparency, re-traceability, re-enaction, re-producibility, explainability
Topic 01 Decison Support Systems (DSS): Can Computers help making better decisions? Students read [1]
Topic 02 History of DSS = History of AI – explainable AI is actually the oldest field of Artificial Intelligence
Topic 03 Medical Informatics Example: Towards P4 Medicine
Topic 04 Medical Informatics Example: Case Based Reasoing (CBR)
Topic 05 Causal Reasoning
Topic 06 Explainability – Causality – Causability Students read [2]
Topic 07 (Some) Methods of Explainable AI
- Lecture slides full size (10.445 kB): 02-185A83-HOLZINGER-EXPLAINABLE-AI-2019ah
- Lecture slides 3 x 3 (8.484 kB): 02-185A83-HOLZINGER-EXPLAINABLE-AI-2019-3x3ah
To get a preview you can have a look at the slides of the last course years: 2018, 2017, 2016
however, please note that for the 2019 exam of course the 2019 slides are relevant
Learning Goals: At the end of this lecture the students …
+ know the roots of decision making and early concepts of medical decision support systems (Advice Taker, MYCIN, GAMUTS)
+ see some examples of the problematic of medical decision making
+ discussion of decsion support of medical experts by AI-systems (also ethical responsibility issues)
+ have a first overview on the principles of explainable AI-methods
+ know some of the most relevant methods of explainable AI
for more details please go to the course page (taking place each semester in Graz):
https://human-centered.ai/explainable-ai-causability-2019
Student read:
[1] Michael Duerr-Specht, Randy Goebel & Andreas Holzinger 2015. Medicine and Health Care as a Data Problem: Will Computers become better medical doctors? In: Holzinger, Andreas, Roecker, Carsten & Ziefle, Martina (eds.) Smart Health, State-of-the-Art SOTA Lecture Notes in Computer Science LNCS 8700. Heidelberg, Berlin, New York: Springer, pp. 21-40, doi:10.1007/978-3-319-16226-3_2.
[Can-Computers-help-doctors-making-better-decisions]
[2] Andreas Holzinger, Georg Langs, Helmut Denk, Kurt Zatloukal & Heimo Mueller 2019. Causability and Explainability of AI in Medicine. Wiley Interdisciplinary Reviews: Data Mining and Knowledge Discovery, doi:10.1002/widm.1312.
Lecture 03 – Week 13, Tuesday, March, 26, 2018
Tutorial T1 & Assignment A1 (Tutor: Anna SARANTI): Layer-wise Relevance Propagation (LRP)
Tutorial T2 & Assignment A2 (Tutor: Marcus BLOICE): Data Discovery and Transfer Learning on Skin Lesion Images
All material can be found on our GitHub page:
https://github.com/human-centered-ai-lab/cla-185A83-machine-learning-health-class-2019
Lecture 04 – Week 14, Tuesday, April, 2, 2019
Module 03 – Probabilistic Graphical Models: From Knowledge Representation to Graph Model Learning
Lecture Outline: In order to get well prepared for the second tutorial on probabilistic programming, this module provides some basics on graphical models and goes towards methods for Monte Carlo sampling from probability distributions based on Markov Chains (MCMC). This is not only very important, it is awesome, as it is similar as our brain may work. It allows for computing hierachical models having a large number of unknown parameters and also works well for rare event sampling wich is often the case in the health informatics domain. So, we start with reasoning under uncertainty, provide some basics on graphical models and go towards graph model learning. One particular MCMC method is the so-called Metropolis-Hastings algorithm which obtains a sequence of random samples from high-dimensional probability distributions -which we are often challenged in the health domain. The algorithm is among the top 10 most important algorithms and is named after Nicholas METROPOLIS (1915-1999) and Wilfred K. HASTINGS (1930-2016); the former found it in 1953 and the latter generalized it in 1970 (remember: Generalization is a grand goal in science).
Lecture Keywords: Reasoning under uncertainty, graph extraction, network medicine, metrics and measures, point-cloud data sets, graphical model learning, MCMC, Metropolis-Hastings Algorithm
Topic 01 Decision Making under uncertainty
Topic 02 Some basics of Markov Processes
Topic 03 A few fundamentals of Concept Learning
Topic 04 Essentials of Graphs/Networks and challenges
Topic 05 Bayes’ Nets
Topic 06 Graphical Model Learning
Topic 07 Probabilistic Programming
Topic 08 Markov Chain Monte Carlo (MCMC)
Topic 09 Example: Metropolis Hastings Algorithm
- Lecture slides full size (8.858 KB): 4-185A83-GRAPHICAL-MODELS-I-HOLZINGER-2019
- Lecture slides 3 x 3 (8.689 KB): 4-185A83-GRAPHICAL-MODELS-I-HOLZINGER-2019-3×3
To get a preview you can have a look at the slides of the last course years: 2018, 2017, 2016
however, please note that for the 2019 exam of course the 2019 slides are relevant
Learning Goals: At the end of this lecture the students
+ are aware of reasoining and decision making
+ have an idea of graphical models
+ understand the advantages of probabilistic programming
Reading for Students:
- Bishop, Christopher M (2007) Pattern Recognition and Machine Learning. Heidelberg: Springer [Chapter 8: Graphical Models]
- Chenney, S. & Forsyth, D. A. 2000. Sampling plausible solutions to multi-body constraint problems. Proceedings of the 27th annual conference on Computer graphics and interactive techniques. ACM. 219-228, doi:10.1145/344779.344882.
- Ghahramani, Z. 2015. Probabilistic machine learning and artificial intelligence. Nature, 521, (7553), 452-459, doi:10.1038/nature14541
- Gordon, A. D., Henzinger, T. A., Nori, A. V. & Rajamani, S. K. Probabilistic programming. Proceedings of the on Future of Software Engineering, 2014. ACM, 167-181, doi:10.1145/2593882.2593900
- KOLLER, Daphne & FRIEDMAN, Nir (2009) Probabilistic graphical models: principles and techniques. Cambridge (MA): MIT press.
- Metropolis, N., Rosenbluth, A. W., Rosenbluth, M. N., Teller, A. H. & Teller, E. 1953. Equation of State Calculations by Fast Computing Machines. The Journal of Chemical Physics, 21, (6), 1087-1092, doi:10.1063/1.1699114. (34,123 citations as of 21.03.2017)
- Wainwright, Martin J. & Jordan, Michael I. (2008) Graphical Models, Exponential Families, and Variational Inference. Foundations and Trends in Machine Learning, Vol.1, 1-2, 1-305, doi: 10.1561/2200000001 [Link to pdf]
- Wood, F., Van De Meent, J.-W. & Mansinghka, V. A New Approach to Probabilistic Programming Inference. AISTATS, 2014. 1024-1032.
A hot topic in ML are graph bandits:
- Villar, S. S., Bowden, J. & Wason, J. 2015. Multi-armed Bandit Models for the Optimal Design of Clinical Trials: Benefits and Challenges. Statistical Science, 199-215, doi:10.1214/14-STS504, accesible via: https://arxiv.org/abs/1507.08025
Lecture 05 – Week 15 – April, 9, 2019
Tutorial T2 – Probabilistic Programming with Python (Tutor: Florian ENDEL) and second assigment
In this tutorial, we will explore probabilistic programming with the Python framework PyMC3. “Probabilistic programming allows for automatic Bayesian inference on user-defined probabilistic models.” [1]
We will start with a brief repetition of the previous lecture by discussing the Bayes’ theorem, Bayesian models and Bayesian parameter estimation using Markov Chain Monte Carlo (MCMC) sampling. Next on, we will dive deeper into the capabilities, workflow and specific utilization of PyMC3. Language primitives, stochastic variables and the intuitive syntax to define complex models and networks will be explored. Increasingly complex examples including, e.g., a simple statistical test, linear (LM) and generalized linear (GLM) models as well as multilevel modelling will highlight the applicability of Bayes’ methodology as well as the potential and simplicity of probabilistic programming with PyMC3. An exercise based on real world research [2] will demonstrate the advantage of multilevel modelling and probabilistic programming.
Introduction to PyMC3: https://florian.endel.at/Presentation/PyMC3Intro/
Assignment Instruction: Exercise-Therapeutic-Touch-LV185A83-2018
The 2019 class will again cover Multilevel Modelling (adapted from Chris Fonnesbeck):
https://florian.endel.at/Presentation/PyMC3Intro/multilevel_modeling#/
Please refer to our Github pages: https://github.com/human-centered-ai-lab/cla-185A83-machine-learning-health-class-2019
[1] John Salvatier, Thomas V. Wiecki & Christopher Fonnesbeck 2016. Probabilistic programming in Python using PyMC3. PeerJ Computer Science, 2, e55, doi:10.7717/peerj-cs.55
[2] Linda Rosa, Emily Rosa, Larry Sarner & Stephen Barrett 1998. A Close Look at Therapeutic Touch. JAMA, 279, (13), 1005-1010, doi:10.1001/jama.279.13.1005
Additional resources:
Lecture slides 2017: full size (815 kB) 2017-04-04 Probabilistic Programming – Endel
Examples 2017: https://github.com/FlorianEndel/Probabilistic-Programming-Tutorial
MCMC: https://chi-feng.github.io/mcmc-demo/app.html
[3] A. Pfeffer, Practical probabilistic programming. Shelter Island, NY: Manning Publications, Co, 2016.
[4] C. Davidson-Pilon, Bayesian methods for hackers: probabilistic programming and Bayesian inference. New York: Addison-Wesley, 2016.
[5] J. K. Kruschke, Doing Bayesian data analysis: a tutorial with R, JAGS, and Stan, Edition 2. Boston: Academic Press, 2015.
Lecture 06 – Week 18, Tuesday, April, 30, 2019
Data for Machine Learning: Quality, fusion, integration, probabilistic information and entropy
Lecture Outline: In this lecture we will review some fundamentals on data and information. In order to carry out successful machine learning, we need not only appropriate algorithms, but above all data. However, it is not only important to have sufficient large amounts of data, but also to have relevant data and the corresponding domain knowledge. You will always get a result, the crucial question is whether and to what extent the results are relevant to support medical decision making from uncertainty; and here we need the concept of Bayes and Laplace and entropy as a measure of uncertainty distributions, and KL divergence as a way of measuring the matching between two distributions.
Lecture Keywords: data, information, probability, entropy, cross-entropy, Kullback-Leibler divergence
Topic 01 Data – The underlying physics of data
Topic 02 Data – Biomedical data sources – taxonomy of data
Topic 03 Data – Integration, Mapping and Fusion of data
Topic 04 Information – Bayes and Laplace probabilistic information p(x)
Topic 05 Information Theory – Information Entropy
Topic 06 Information Cross-Entropy and Kullback-Leibler Divergence
To get a preview you can have a look at the slides of the last course years: 2018, 2017, 2016
however, please note that for the 2019 exam of course the 2019 slides are relevant
Learning Goals: At the end of this lecture the students
+ are aware of the problematic of health data and understand the importance of data integration in the life sciences.
+ understand the concept of probabilistic information with a focus on the problem of estimating the parameters of a Gaussian distribution (maximum likelihood problem).
+ recognize the usefulness of the relative entropy, called Kullback–Leibler divergence which is very important, particularly for sparse variational methods between stochastic processes.
Reading for Students: (some prereading/postreading recommendations):
- Banerjee, O., El Ghaoui, L. & D’aspremont, A. 2008. Model selection through sparse maximum likelihood estimation for multivariate gaussian or binary data. The Journal of Machine Learning Research, 9, 485-516, https://www.jmlr.org/papers/v9/banerjee08a.html
- De Boer, P.-T., Kroese, D. P., Mannor, S. & Rubinstein, R. Y. 2005. A tutorial on the cross-entropy method. Annals of operations research, 134, (1), 19-67. doi:10.1007/s10479-005-5724-z
- Galas, D. J., Dewey, T. G., Kunert-Graf, J. & Sakhanenko, N. A. 2017. Expansion of the Kullback-Leibler Divergence, and a new class of information metrics. arXiv:1702.00033.
- Holzinger, A., Dehmer, M. & Jurisica, I. (2014). Knowledge Discovery and interactive Data Mining in Bioinformatics – State-of-the-Art, future challenges and research directions. BMC Bioinformatics, 15, (S6), I1. doi:10.1186/1471-2105-15-S6-I1
- Holzinger, A., Hörtenhuber, M., Mayer, C., Bachler, M., Wassertheurer, S., Pinho, A. & Koslicki, D. (2014). On Entropy-Based Data Mining. In: Holzinger, A. & Jurisica, I. (eds.) Interactive Knowledge Discovery and Data Mining in Biomedical Informatics, Lecture Notes in Computer Science, LNCS 8401. Berlin Heidelberg: Springer, pp. 209-226. doi:10.1007/978-3-662-43968-5_12
Online available: https://pure.tugraz.at/portal/files/3108669/HOLZINGER_Entropy_based_data_mining.pdf - Loshchilov, Ilya, Schoenauer, Marc & Sebag, Michele (2013). KL-based Control of the Learning Schedule for Surrogate Black-Box Optimization. arXiv:1308.2655.
- Matthews, A., Hensman, J., Turner, R. E. & Ghahramani, Z. On sparse variational methods and the Kullback-Leibler divergence between stochastic processes. Proceedings of the Nineteenth International Conference on Artificial Intelligence and Statistics (AISTATS), 2016. JMLR, 231-239 https://www.jmlr.org/proceedings/papers/v51/matthews16.html
Additional Reading: (to foster a deeper understanding of information theory related to the life sciences):
- Manca, Vincenzo (2013). Infobiotics: Information in Biotic Systems. Heidelberg: Springer. (This book is a fascinating journey through the world of discrete biomathematics and a continuation of the 1944 Paper by Erwin Schrödinger: What Is Life? The Physical Aspect of the Living Cell, Dublin, Dublin Institute for Advanced Studies at Trinity College)
Lecture 07, Tuesday, May, 7, 2019
Module 05 – Causality, Explainability, Ethical, Legal and Social issues of AI/ML in health informatics
Keywords: Causality, Graphical Causal Models, AI Ethics
Topic 01: Causality
Topic 02: Explainability and Causability
Topic 03: AI Ethics
Topic 04: Social Issues of AI
- Lecture slides full size (4.746 KB): 07-185A83-HOLZINGER-CAUSALITY-AI-ETHICS-2019
- Lecture slides 3 x 3 (1.924 KB): 07-185A83-HOLZINGER-CAUSALITY-AI-ETHICS-2019-3×3
To get a preview you can have a look at the slides of the last course years: 2018, 2017, 2016
however, please note that for the 2019 exam of course the 2019 slides are relevant
Learning Goals: At the end of this lecture the students …
+ have a basic overview on the problem of causality, causal inference and functional causal models
+ can compare the issues of causality with issues of usability
+ have aquired some undertanding of reasoning (deductive, inductive, abductive)
+ are aware of the difficulty of hard inference problems
+ have a feeling on the problems of AI Ethics, laws and social issues of AI
Reading for students:
[0] Jonas Peters, Dominik Janzing & Bernhard Schölkopf 2017. Elements of causal inference: foundations and learning algorithms, Cambridge (MA), online available at:
https://web.math.ku.dk/~peters/elements.html
[1] Judea Pearl 1988. Evidential reasoning under uncertainty. In: Shrobe, Howard E. (ed.) Exploring artificial intelligence. San Mateo (CA): Morgan Kaufmann, pp. 381-418.
[2] Matt J. Kusner, Joshua Loftus, Chris Russell & Ricardo Silva. Counterfactual fairness. In: Guyon, Isabelle, Luxburg, Ulrike Von, Bengio, Samy, Wallach, Hanna, Fergus, Rob & Vishwanathan, S.V.N., eds. Advances in Neural Information Processing Systems 30 (NIPS 2017), 2017. 4066-4076.
[3] Judea Pearl 2009. Causality: Models, Reasoning, and Inference (2nd Edition), Cambridge, Cambridge University Press.
[4] Judea Pearl 2018. Theoretical Impediments to Machine Learning With Seven Sparks from the Causal Revolution. arXiv:1801.04016.
Final Lecture 08, Tuesday, May, 28, 2019
The grading consists of three independent parts:
I) Final Exam (written test quiz, 30%) – see sample exam here
STUDENT-LV-185A83-Machine-Learning-for-Health-Informatics-Exam-class-of-2019
II) Presentations of the assigments (orally, 10 %)
III) Grading of the assignments (coding, 20 % each, 60 % total)
Note: The course will be adpated to the students accordingly as the course progresses. Each lecture is preceded by a quiz from the last lecture. The slides will be put online AFTER each lecture – and only those are binding for the final exam. Note that the slides presented and the slides showed on the Web can be different for didactical purposes.
Short Bio of Lecturer:
Andreas HOLZINGER <expertise> promotes a synergistic approach to Human-Centred AI (HCAI) and has pioneered in interactive machine learning (iML) with the human-in-the-loop. He promotes an integrated machine learning approach with the goal to augment human intelligence with artificial intelligence to help to solve problems in health informatics.
Due to raising ethical, social and legal issues governed by the European Union, future AI supported systems must be made transparent, re-traceable, thus human interpretable. Andreas’ aim is to explain why a machine decision has been reached, paving the way towards explainable AI and Causability, ultimately fostering ethical responsible machine learning, trust and acceptance for AI.
Andreas obtained a Ph.D. in Cognitive Science from Graz University in 1998 and his Habilitation (second Ph.D.) in Computer Science from Graz University of Technology in 2003. Andreas was Visiting Professor for Machine Learning & Knowledge Extraction in Verona, RWTH Aachen, University College London and Middlesex University London. Since 2016 Andreas is Visiting Professor for Machine Learning in Health Informatics at the Faculty of Informatics at Vienna University of Technology. Currently, Andreas is Visiting Professor for explainable AI, Alberta Machine Intelligence Institute, University of Alberta, Canada.
Group Homepage: https://explainable-ai.org
Personal Homepage: https://www.aholzinger.at
Youtube Introduction Video: https://youtu.be/lc2hvuh0FwQ
Conference Homepage: https://cd-make.net
Short Bio of Tutors:
Marcus BLOICE is finishing his PhD this year with the application of deep learning on medical images. Currently, he is working on the Augmentor project and the Digital Pathology project, and is involved in the featureCloud project. He has a background in computer science from the University of Sunderland (UK). He is a programmer in Python and has experience with the popular machine learning pipelines. Marcus has also experience in machie learning on large medical images.
Florian ENDEL started working as a database developer in the general field of healthcare research in 2007 – after gathering first experiences as high school teacher for two years and working as freelance Web designer, A specific highlight is the development and supervision of “GAP-DRG”, a database holding massive amounts of reimbursement data from the Austrian social insurance system, since 2008. Since then, he was part of several national and international research projects handling, among others, data management, data governance, statistical analytics and secure computing infrastructure. He is currently participating in the EU FP7 project CEPHOS-LINK, the FFG K-Projekt DEXHELPP and still finishing his master’s thesis.
Anna SARANTI is just finalizing her Master’s studies with a work on Applying Probabilistic Graphical Models and Deep Reinforcement Learning in a Learning-Aware Application, supervised by Andreas Holzinger and Martin Ebner at Graz University of Technology. Anna is currently working as machine learning engieer in Vienna.
Additional pointers and reading suggestions can be found a the
Learning Machine Learning page
Excellent Ressources for excercises
Github repository by Alberto Blanco Garcés https://github.com/alberduris
Related Books in Machine Learning:
- MITCHELL, Tom M., 1997. Machine learning, New York: McGraw Hill. (Book Webpages)
Undoubtedly, this is the classic source from the pioneer of ML for getting a perfect first contact with the fascinating field of ML, for undergraduate and graduate students, and for developers and researchers. No previous background in artificial intelligence or statistics is required. - FLACH, Peter, 2012. Machine Learning: The Art and Science of Algorithms that Make Sense of Data. Cambridge: Cambridge University Press. (Book Webpages)
Introductory for advanced undergraduate or graduate students, at the same time aiming at interested academics and professionals with a background in neighbouring disciplines. It includes necessary mathematical details, but emphasizes on how-to. - MURPHY, Kevin, 2012. Machine learning: a probabilistic perspective. Cambridge (MA): MIT Press. (Book Webpages)
This books focuses on probability, which can be applied to any problem involving uncertainty – which is highly the case in medical informatics! This book is suitable for advanced undergraduate or graduate students and needs some mathematical background. - BISHOP, Christopher M., 2006. Pattern Recognition and Machine Learning. New York: Springer-Verlag. (Book Webpages)
This is a classic work and is aimed at advanced students and PhD students, researchers and practitioners, not asuming much mathematical knowledge. - HASTIE, Trevor, TIBSHIRANI, Robert, FRIEDMAN, Jerome, 2009. The Elements of Statistical Learning: Data Mining, Inference, and Prediction. New York: Springer-Verlag (Book Webpages)
This is the classic groundwork from supervised to unsupervised learning, with many applications in medicine, biology, finance, and marketing. For advanced undergraduates and graduates with some mathematical interest.
To get an understanding of the complexity of the health informatics domain:
- Andreas HOLZINGER, 2014. Biomedical Informatics: Discovering Knowledge in Big Data.
New York: Springer. (Book Webpage)
This is a students textbook for undergraduates, and graduate students in health informatics, biomedical engineering, telematics or software engineering with an interest in knowledge discovery. This book fosters an integrated approach, i.e. in the health sciences, a comprehensive and overarching overview of the data science ecosystem and knowledge discovery pipeline is essential. - Gregory A PETSKO & Dagmar RINGE, 2009. Protein Structure and Function (Primers in Biology). Oxford: Oxford University Press (Book Webpage)
This is a comprehensive introduction into the building blocks of life, a beautiful book without ballast. It starts with the consideration of the link between protein sequence and structure, and continues to explore the structural basis of protein functions and how this functions are controlled. - Ingvar EIDHAMMER, Inge JONASSEN, William R TAYLOR, 2004. Protein Bioinformatics: An Algorithmic Approach to Sequence and Structure Analysis. Chicheser: Wiley.
Bioinformatics is the study of biological information and biological systems – such as of the relationships between the sequence, structure and function of genes and proteins. The subject has seen tremendous development in recent years, and there are ever-increasing needs for good understanding of quantitative methods in the study of proteins. This book takes the novel approach of covering both the sequence and structure analysis of proteins and from an algorithmic perspective.
Amongst the many tools (we will concentrate on Python), some useful and popular ones include:
- WEKA. Since 1993, the Waikato Environment for Knowledge Analysis is a very popular open source tool. In 2005 Weka received the SIGKDD Data Mining and Knowledge Discovery Service Award: it is easy to learn and easy to use [WEKA]
- Mathematica. Since 1988 a commercial symbolic mathematical computation system, easy to use [Mathematica]
- MATLAB. Short for MATrix LABoratory, it is a commercial numerical computing environment since 1984, coming with a proprietary programming language by MathWorks, very popular at Universities where it is licensed, awkward for daily practice [Matlab]
- R. Coming from the statistics community it is a very powerful tool implementing the S programming language, used by data scientists and analysts. [The R-Project]
- Python. Currently maybe the most popular scientific language for ML [Python Software Foundation]
An excellent source for learning numerics and science with Python is: https://www.scipy-lectures.org/ - Julia. Since 2012, raising scientific language for technical computing with better performance than Python. IJulia, a collaboration between the Jupyter and Julia, provides a powerful browser-based graphical notebook interface to Julia. [julialang.org]
Please have a look at: What tools do people generally use to solve problems?
Recommendable reading on tools include:
- Wes McKINNEY (2012) Python for Data Analysis: Data Wrangling with Pandas, NumPy, and IPython. Beijing et al.: O’Reilly.
This is a practical introduction from the author of the Pandas library. [Google-Books] - Ivo BALBAERT (2015) Getting Started with Julia Programming. Birmingham: Packt Publishing.
A good start for the Julia language and more focused on scientific computing projects, it is assumed that you already know about a high-level dynamic language such as Python. [Google-Books]
International Courses on Machine Learning:
- Carnegie Mellon University > Machine Learning Course 10-701 2015
by Eric XING (expertise) and Ziv-Bar JOSEPH (expertise)
https://www.cs.cmu.edu/~epxing/Class/10701/lecture.html - Carnegie Mellon University > Machine Learning Course 10-701/15-781 2011
by Tom MITCHELL (expertise)
https://www.cs.cmu.edu/~tom/10701_sp11/ - Carnegie Mellon University > Machine Learning Course 10-601 2015
by Maria-Florina BALCAN (expertise) and Tom MITCHELL (expertise)
https://www.cs.cmu.edu/~ninamf/courses/601sp15/ - Carnegie Mellon University > Machine Learning Course 10-701 2013
by Alex SMOLA (expertise)
https://alex.smola.org/teaching/cmu2013-10-701/index.html - Carnegie Mellon University > Machine Learnigng Course 10601b 2015
by Seyoung KIMhttps://www.cs.cmu.edu/~10601b/ - Cornell University > Machine Learning CS 4780/5780 2014
by Thorsten JOACHIMS (expertise)
https://www.cs.cornell.edu/courses/cs4780/2014fa/ - Cornell University > General Machine Learning, Knowledge Extraction
and Data Science courses
https://machinelearning.cis.cornell.edu/pages/courses.php - Oxford > Department of Computer Science > Machine Learning: 2014-2015
by Nando de FREITAS (expertise)
https://www.cs.ox.ac.uk/teaching/courses/2014-2015/ml/index.html
Conferences on Machine Learning with a special focus on health application
- CD-MAKE – Cross Domain Conference for MAchine Learning and Knowledge Extraction
https://cd-make.net
- NIPS (now called NeurIPS) – has always workshops on machine learning for health
https://neurips.cc - ICML – has also always workshops/sessions on and for health
https://icml.cc/
Pointers:
A) Students with a GENERAL interest in machine learning should definitely browse these sources:
- TALKING MACHINES – Human conversation about machine learning by Katherine GORMAN and Ryan P. ADAMS <expertise>
excellent audio material – 24 episodes in 2015 and three new episodes in season two 2016 (as of 14.02.2016) - This Week in Machine Learning and Artificial Intelligence Podcast
https://twimlai.com - Data Skeptic – Data science, statistics, machine learning, artificial intelligence, and scientific skepticism
https://dataskeptic.com - VIDEOLECTURES.NET Machine learning talks (3,580 items up to 31.01.2017) ML is grouped into subtopics
and displayed as map – highly recommendable - TUTORIALS ON TOPICS IN MACHINE LEARNING by Bob Fisher from the University of Edinburgh, UK
B) Students with a SPECIFIC interest in interactive machine learning should have a look at:
https://human-centered.ai/lv-706-315-interactive-machine-learning/